GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.
Python package for analysis of multiomic single cell RNA-seq and ATAC-seq. - GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells

Integrative analyses of single-cell transcriptome and regulome using MAESTRO, Genome Biology

CSS: cluster similarity spectrum integration of single-cell genomics data, Genome Biology
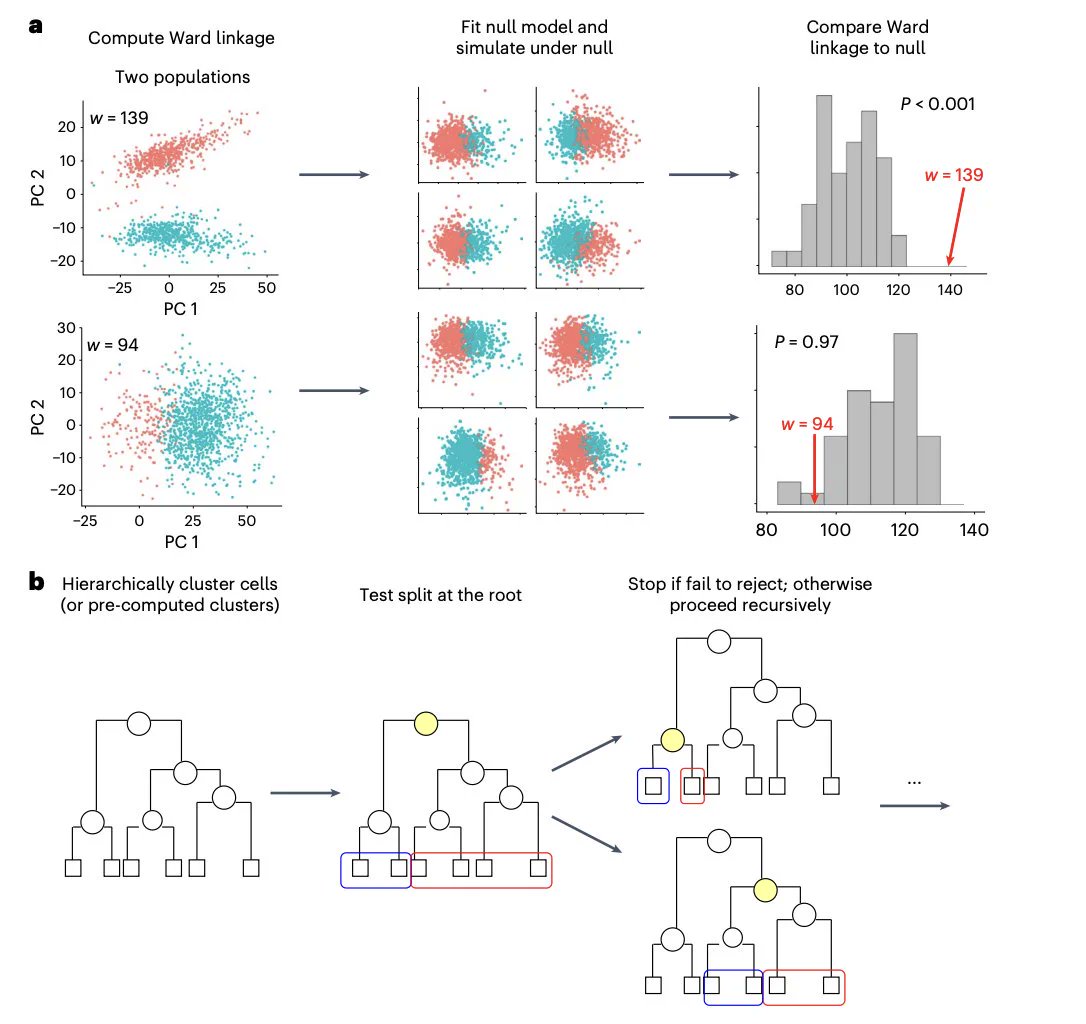
CFCE-DFCI (@CFCE_DanaFarber) / X

Multi-omic single-cell velocity models epigenome–transcriptome interactions and improves cell fate prediction

Integrated analysis of multimodal single-cell data - ScienceDirect
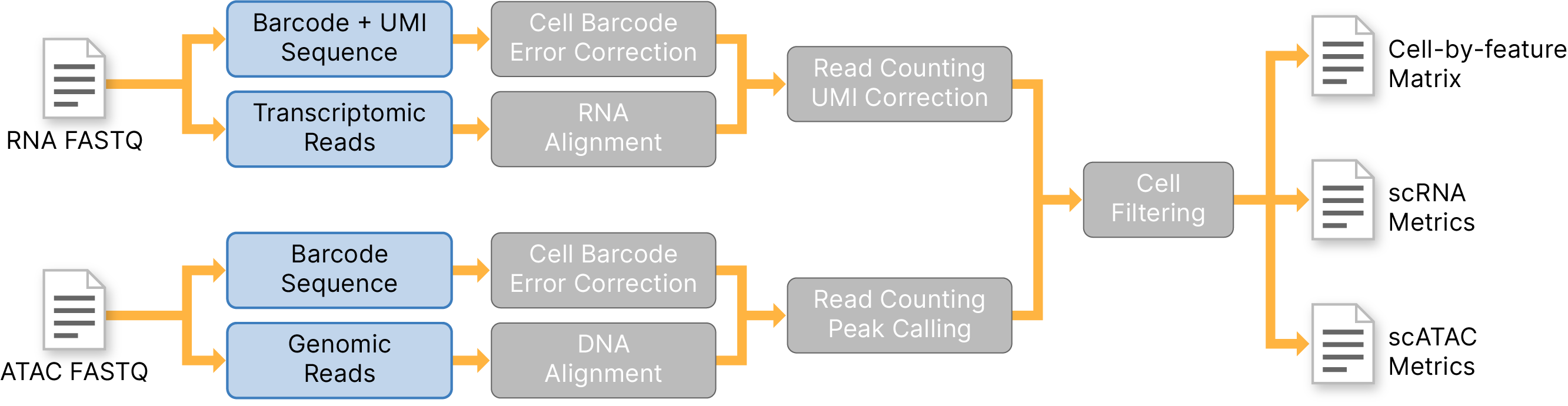
Multiomics Pipeline
GitHub - quadbio/scMultiome_analysis_vignette: Tutorial of single-cell RNA-ATAC multiomic sequencing data analysis
GitHub - zhongmicai/awesome_single_cell

Integrating single-cell transcriptomes, chromatin accessibility, and multiomics analysis of mesoderm-induced embryonic stem cells - ScienceDirect

Nonparametric single-cell multiomic characterization of trio relationships between transcription factors, target genes, and cis-regulatory regions - ScienceDirect
GitHub - mdozmorov/RNA-seq_notes: A continually expanding collection of RNA- seq tools
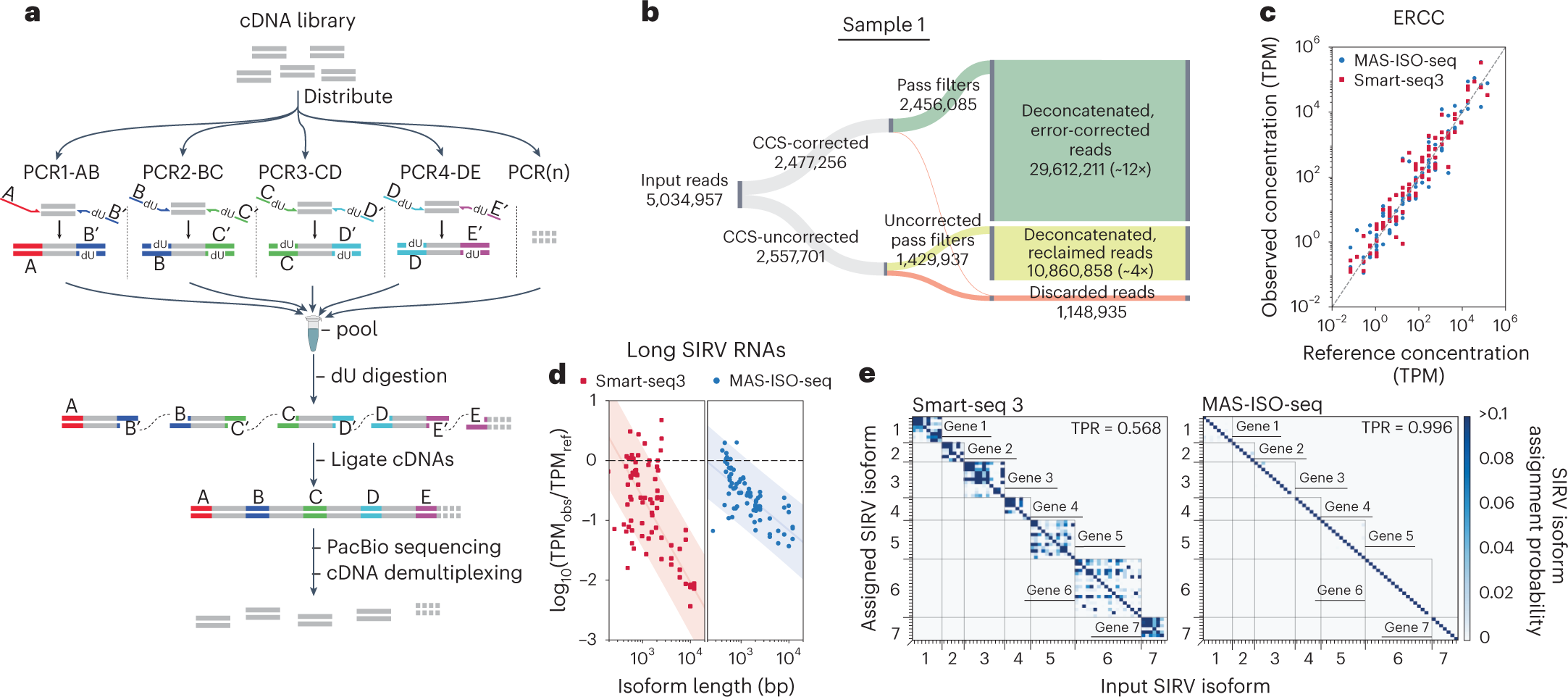
High-throughput RNA isoform sequencing using programmed cDNA concatenation

A multi-omics atlas of the human retina at single-cell resolution - ScienceDirect









