Principle of SNAP/CLIP-labeling and labeling protocols. (a) SNAP-and
Download scientific diagram | Principle of SNAP/CLIP-labeling and labeling protocols. (a) SNAP-and CLIP-tag fused to the protein of interest bind benzylguanine-or benzylcytosine-substrates, respectively. There are various cell-permeable fluorescent substrates and a non-fluorescent substrate available. (b) For labeling of the whole fusion protein pool cells are incubated with a fluorescent substrate, washed and fixed/imaged. (c) For age-dependent labelings cells are first incubated with the non-fluorescent blocker followed by labeling of the newly-synthesized fusion protein with a fluorescent substrate. To omit continuous labeling with the first substrate additional blocking/ labeling steps can be added. from publication: A Global Approach for Quantitative Super Resolution and Electron Microscopy on Cryo and Epoxy Sections Using Self-labeling Protein Tags | Correlative light and electron microscopy (CLEM) is a powerful approach to investigate the molecular ultrastructure of labeled cell compartments. However, quantitative CLEM studies are rare, mainly due to small sample sizes and the sensitivity of fluorescent proteins to | Protein Tagging, Electron Microscopy and Correlative Light Electron Microscopy | ResearchGate, the professional network for scientists.

Halo SNAP CLIP for Protein Labeling - Bitesize Bio

Principle of SNAP/CLIP-labeling and labeling protocols. (a) SNAP-and

HTRF Tag-lite SNAP-Red labeling reagent

Comparison of Halo and SNAP Tagging in Live-Cell Super-Resolution
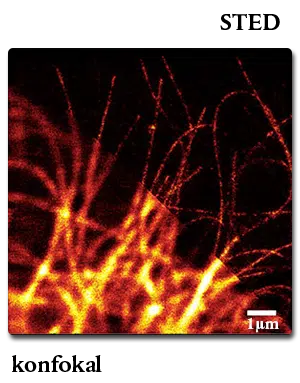
Cellular Analysis (SNAP-Tag) - New England Biolabs GmbH
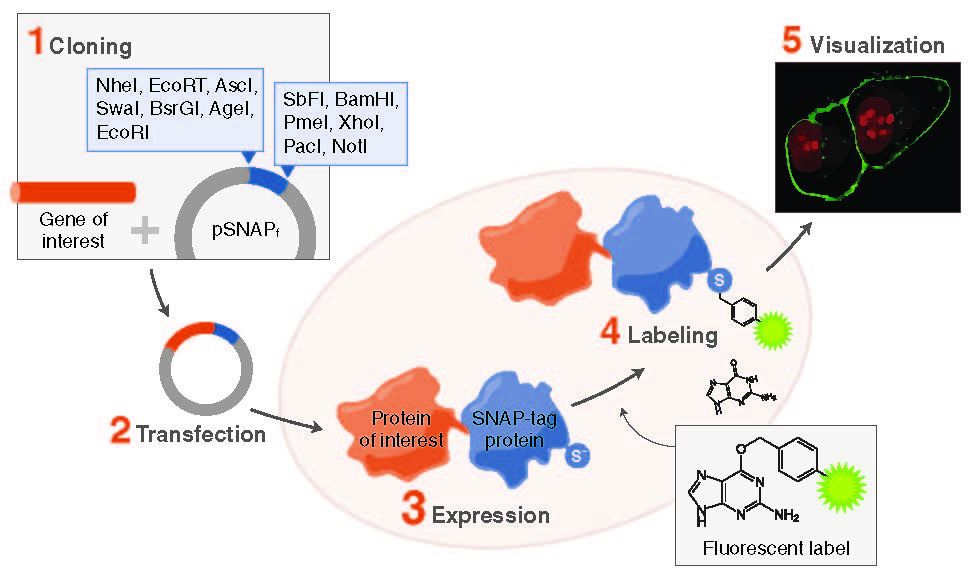
SNAP-tag® Technologies: Tools to Study Protein Function
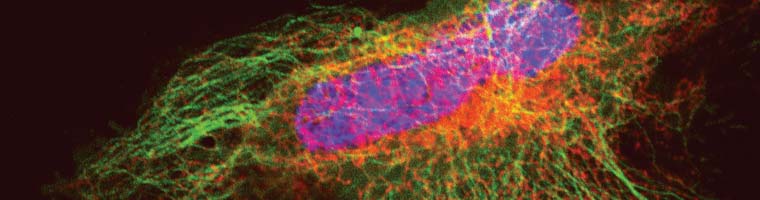
Cellular Analysis (SNAP-Tag) - New England Biolabs GmbH

1 Three general strategies when considering CLEM in experimental

Comparison of HaloTag and SNAP-Tag Labeling with Various Fluorophores

Labeling and Single-Molecule Methods To Monitor G Protein-Coupled Receptor Dynamics

Jean-Marc VERBAVATZ, Group Leader, Institut Jacques Monod, Paris, IJM
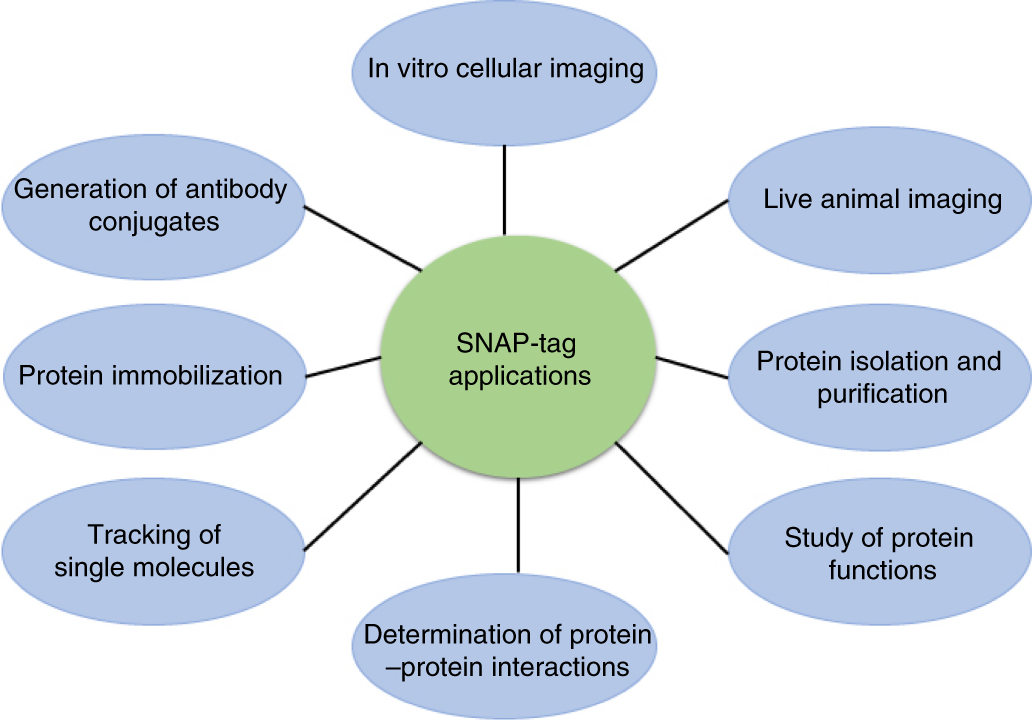
One-step site-specific antibody fragment auto-conjugation using SNAP-tag technology

A Global Approach for Quantitative Super Resolution and Electron Microscopy on Cryo and Epoxy Sections Using Self-labeling Protein Tags