GitHub - schatzlab/crossstitch: Code for phasing SVs with SNPs
Code for phasing SVs with SNPs. Contribute to schatzlab/crossstitch development by creating an account on GitHub.
chipimputation/docs/Tutorial.md at master · h3abionet/chipimputation · GitHub
GitHub - schatzlab/crossstitch: Code for phasing SVs with SNPs
phasing · GitHub Topics · GitHub

Automated assembly of high-quality diploid human reference genomes
GitHub - hennlab/snake-SNP_QC: Henn Lab SNP Array QC pipeline from GenomeStudio Output

Automated assembly of high-quality diploid human reference genomes

Semi-automated assembly of high-quality diploid human reference genomes. - Abstract - Europe PMC
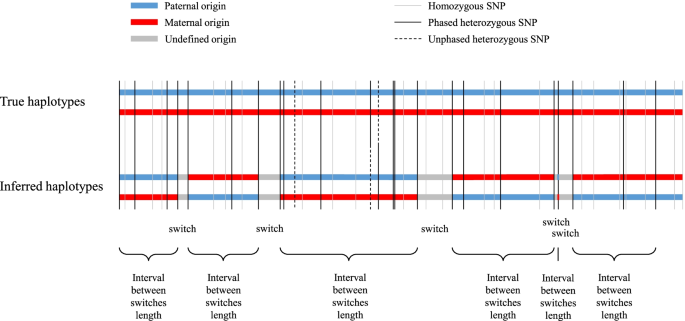
Benchmarking phasing software with a whole-genome sequenced cattle pedigree, BMC Genomics
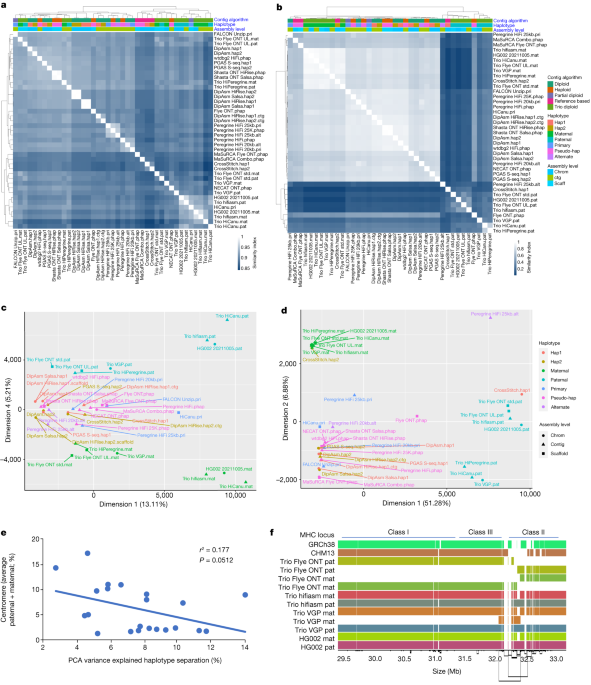
Semi-automated assembly of high-quality diploid human reference genomes
security/xss.md at master · shivapbhusal/security · GitHub

How to add comparissons between treatments for heach gene expression in the same plot? ggboxplot · Issue #169 · kassambara/ggpubr · GitHub

GitHub - apriha/snps: tools for reading, writing, merging, and remapping SNPs
Heterozygous SVs and homozygous SVs · Issue #72 · fritzsedlazeck/Sniffles · GitHub
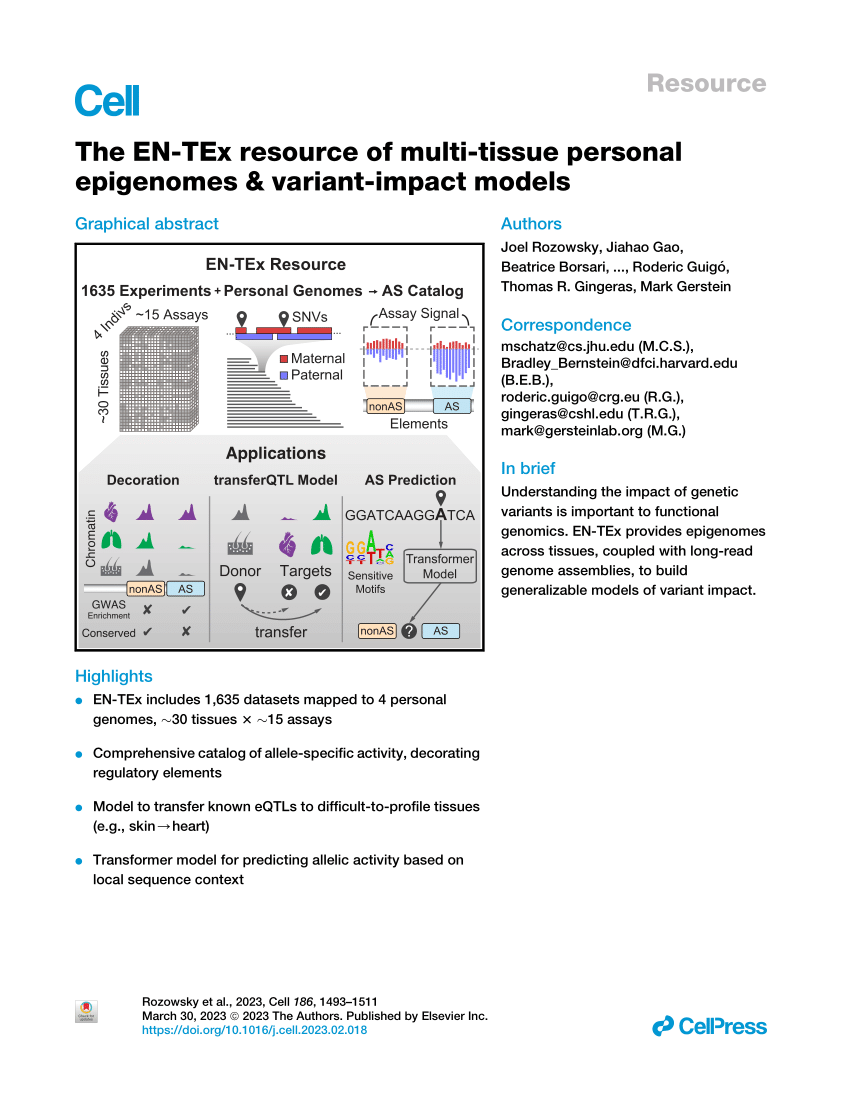
PDF) The EN-TEx resource of multi-tissue personal epigenomes & variant-impact models









